Tokyo Tech News
Tokyo Tech News
Published: March 31, 2011
Tokyo Institute of Technology researchers reveal the molecular mechanisms through which bacterial biofilms depend on the presence of blue light
In the microscopic world, the saying ‘strength in numbers’ is particularly true. Micro-organisms such as bacteria often gather close together, and produce extracellular proteins to form a protective slime around the colony. This aggregate of cells is called a biofilm and it is the most common form of microbial life in the natural world, providing not only protection from external stresses but also improved pathways for gene transfer.
Unfortunately for humans, biofilms are believed to be involved in the majority of microbial infections. Therefore biochemists hope to uncover the environmental and molecular factors that influence biofilm formation.
For example, studies have revealed that one particular environmental factor - the presence of blue light – is important for biofilm formation in colonies of the intestinal bacterium Escherichia coli, the freshwater bacterium Caulobacter crescentus and the deep-sea bacterium Idiomarina loihiensis. Although these three organisms live in very different environments, they have one thing in common: they do not perform photosynthesis.
Now, Tokyo Institute of Technology researchers have contributed to research which reveals that blue light is also required for biofilm formation in Rhodopseudomonas palustris, a purple photosynthetic bacterium found in earthworm droppings, marine sediments and pond water. More importantly the researchers, led by Shinji Masuda at the Center for Biological Resources and Informatics1, have identified the molecular signalling pathways that produce this link.
A key role in biofilm formation is played by a messenger protein called cyclic dimeric GMP (c-di-GMP), which is ubiquitously present in bacteria and contributes to several other processes including cell motility, virulence and the cell cycle. c-di-GMP is degraded (hydrolyzed) by enzymes called phosphodiesterases, which include a specialized structural domain called EAL.
Another important family of proteins contains the BLUF domain, a photoreceptor sensitive to blue light. BLUF has been shown to modulate many physiological activities such as the expression of photosynthesis genes in response to differing light levels. However, until recently it has been unclear exactly how signals are transferred from BLUF to other domains such as EAL.
Masuda and co-workers focussed on the interactions between a BLUF protein called PapB and an EAL protein called PapA, which are both found in R. palustris. To do this, they grew wild-type R. palustris alongside three mutant strains: one with the PapB gene knocked out, one with PapA knocked out, and one with both PapA and PapB knocked out. All the cultures were grown for 70 hours, with some samples in the dark and some under blue light.
The researchers found that no significant biofilms formed in any of the samples grown in the dark. Where blue light was present, biofilms were seen in all the samples, however there were far more extensive biofilms in the mutant strains where PapB was knocked out.
As the researchers report in their paper, published in the journal Biochemistry: “In response to blue light, PapB increased the c-di-GMP hydrolysis activity of PapA, which inhibited biofilm formation.”
Interestingly, the PapA mutant also showed increased biofilm formation relative to the wild type control, but not to the same extent as the PapB mutant. This implies that PapB may also interact with some other EAL proteins to reduce the cellular levels of c-di-GMP. As the researchers point out, this is highly likely because the R. palustris genome contains more than fifteen genes that encode enzymes with EAL domains.
As a final step, the researchers ran computer simulations of the molecular dynamics in the system, revealing the detailed structural changes that likely occur when PapB binds to the EAL domain on PapA. Their research represents the first full illustration of how a blue-light signal is transferred downstream to influence a major physiological process.
In fact, Masuda and co-workers hope their research might allow scientists to deliberately control processes such as biofilm formation. As they conclude: “A more detailed understanding of the PapA-PapB interaction might find use in a method that allows the artificial control of cellular c-di-GMP levels by BLUF proteins.”
1.T. Kanazawa, et al. Biochemical and physiological characterization of a BLUF protein—EAL protein complex involved in blue light-dependent degradation of cyclic diguanylate in the purple bacterium Rhodopseudomonas palustris.
Biochemistry 49, 10647–10655 (2010).
R. palustirs grown photosynthetically
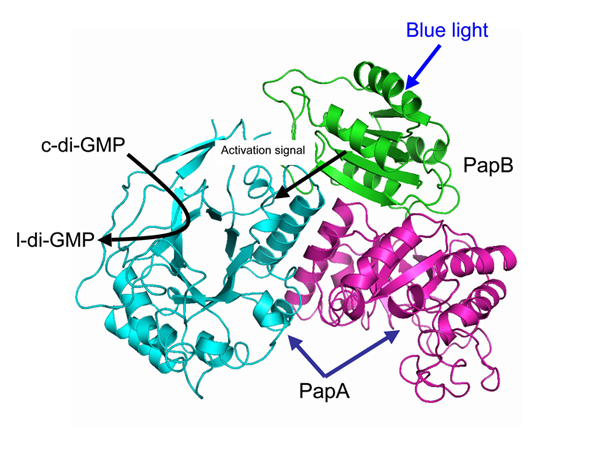
The structure model of the PapA-PapB complex
Reference
Shinji Masuda
Center for Biological Resources and Informatics
Associate Professor