A simple genetic circuit developed at Tokyo Institute of Technology could allow researchers to control how cells diversify into different functional types
During the development of organisms, cell populations diversify into groups with distinct physical or functional characteristics, called phenotypes. This diversification is also important for regeneration after tissue damage, and is driven by complex interactions between different genes. While lots might be known about individual genes, the precise interactions that determine phenotypes are difficult to verify. This provides strong motivation to develop simple, synthetic genetic circuits that emulate natural phenomena while being easier to interpret.
Now, Daisuke Kiga and co-workers from Tokyo Institute of Technology and the University of Tokyo have constructed a simple circuit of just four genes, which illustrates how diversification is influenced by signalling between different cells1. Their work could potentially be applied in tissue engineering to encourage the preferential development of certain cell types for medical purposes.
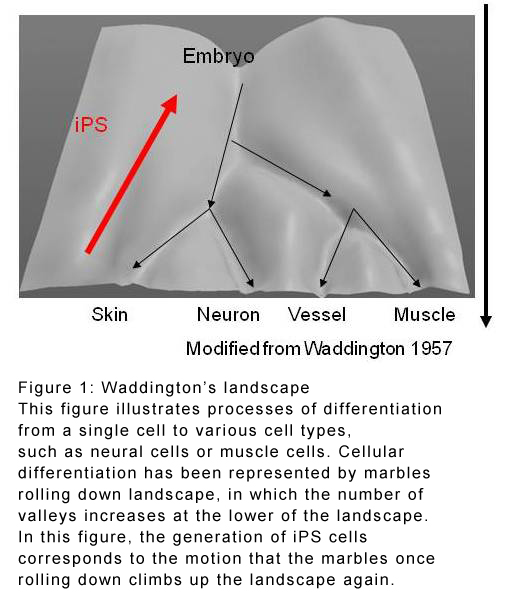
A useful concept for understanding cell diversification is 'Waddington's landscape': a gentle U-shaped slope that splits into different valleys on moving downhill. Cells are imagined as marbles that tumble down the landscape, ending up in a valley representing a certain phenotype. Previous work has shown that phenotype changes, represented by splitting valleys, result not just from the passage of time but also from changes in genetic expression in individual cells. There is also much evidence that signalling between cells holds a strong influence over diversification.
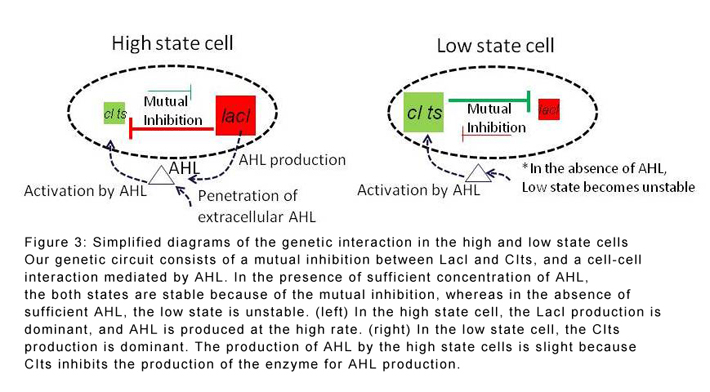
With this in mind, Kiga and co-workers developed a simple genetic circuit with E. coli populations. The four genes in the circuit operate a 'toggle switch' that transforms cells from one state to another, and an intercellular signalling system based on a signalling molecule called acyl-homoserine lactone (AHL).
The study was successful because the researchers integrated both experiments and simulations. "Simulation is a fast and efficient tool for determining how to progress the experiments", says Kiga. "Indeed, we improved experimental conditions by reference to our simulation."
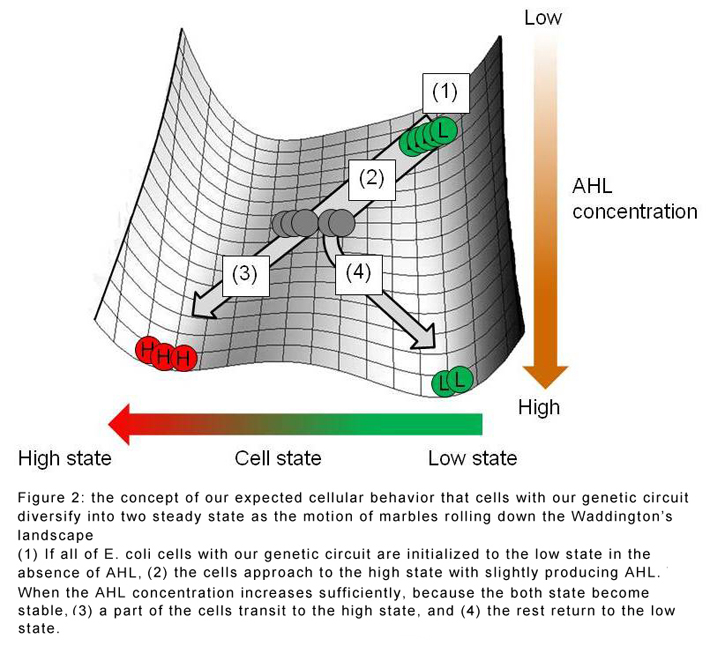
The researchers found that the population tended to split into two distinct phenotypes: a 'high' state where AHL is produced at a rapid rate, and a 'low' state where AHL production is suppressed. The balance in numbers between the two states depends strongly on the speed at which AHL is accumulating in the system. This means that if all the cells start in the low state, cells will diversify to the high state until they are producing enough AHL to support a mixed population of both types of cell.
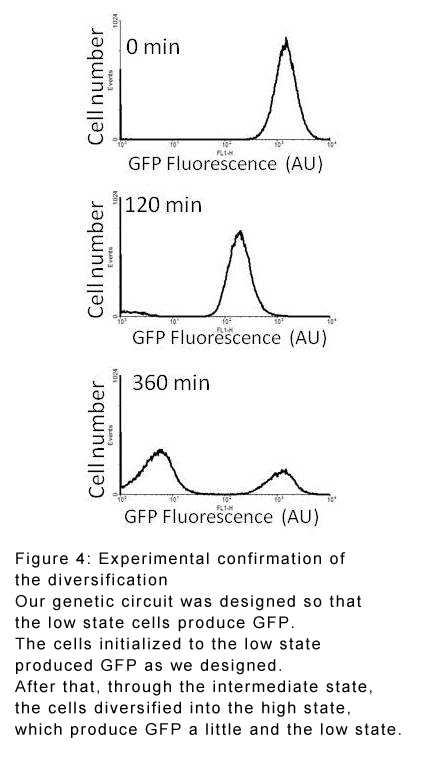
These findings were confirmed in E. coli culture experiments in which the low state was modified to give off green fluorescence whereas the high state did not. In the initial low-state-only population, only one fluorescence peak was seen. After 3 hours, the peak had shifted and widened, and after around 6 hours there were two clearly separate peaks representing both high and low state cells.
This work, and a stochastic model of the genetic circuit, showed that the speed of diversification depended not only on the AHL production rate of individual cells, but also the overall cell density in the population. This finding is reminiscent of the 'community effect', whereby the fates of cells are strongly affected by their neighbours.
In this case, the community effect probably arises because intercellular signalling via AHL is more successful when cells are packed closer together. Such findings could provide a model for understanding development in higher organisms; indeed some other studies have indicated that the differentiation of embryonic stem cells is dependent on cell numbers.
The researchers are also hopeful that simple genetic circuits like theirs could provide useful tools for tissue engineering. These tools could work by providing control over the ultimate fate of cells during development of tissues for specific medical purposes.
"We plan to extend our synthetic circuit from bacterial cells to human cells," explains Kiga. "The circuit will be used to create new feeder cells, rather than to directly induce stem cells. Because our circuit functioning depends on the initial cell density, the resulting cell types can be regulated by changing the density of the feeder cells."
- 1.
- Sekine, R. et al. (2011) Tunable synthetic phenotypic diversification on Waddington's landscape through autonomous signalling. PNAS, doi: 10.1073/pnas.1105901108
. Any information published on this site will be valid in relation to Science Tokyo.






