Summary
The Tokyo Institute of Technology research group of Kentaro Ito (graduate student, Graduate School of Bioscience and Biotechnology), Yumiko Kawano (former graduate student), Yasuhiro Tsutsui (Assistant Professor), Hiroshi Iwasaki (Professor), and Yumiko Kurokawa (Assistant Professor by special-appointment, Academy of Computational Life Sciences) et al. succeeded in elucidating the complicated regulating mechanism in homologous recombination *1. The group's findings are expected to contribute not only to fundamental biology but also to medical research in carcinogenic mechanisms and premature aging.
In particular, the group's research ascertained that Fbh1 *2, which has two different enzymatic activities, qualitatively and quantitatively regulates Rad51 recombinase *3, an enzyme that plays a central role in homologous recombination. Essentially, the group's research indicated that (i) DNA helicase activity *4 of Fbh1 facilitates proper recombination reactions of Rad51 and inhibits improper recombination reactions, and (ii) ubiquitin ligase activity *5 facilitates the ubiquitination of Rad51. The mechanism of homologous recombination in mitosis works mainly to repair DNA double-strand breaks (recombination repair) and contributes genome stability. If for some reason, however, improper chromosome DNA rearrangement occurs due to the reckless recombination, this may result in oncogenic transformation or cell death. Therefore, recombination is tightly regulated in various steps to ensure it proceeds appropriately.
This research was a collaboration with the International Institute for Advanced Studies. The results were published online in PLoS Genetics, an open access journal, on August 28, 2014.
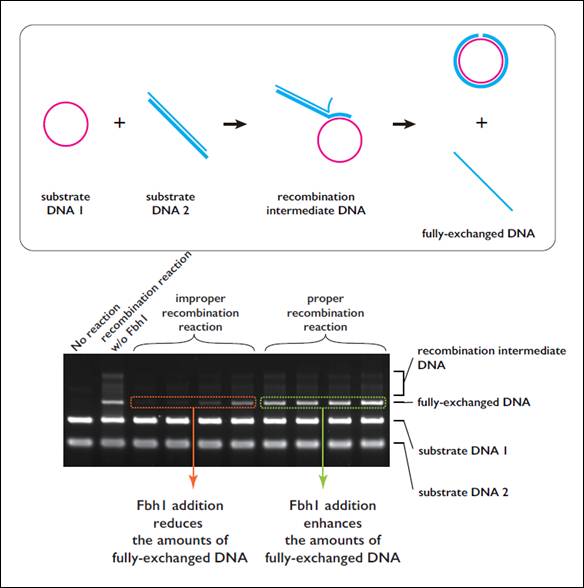
Figure 1. Fbh1 qualitatively regulates the recombination reaction
(Area within frame) Schematic diagram of a recombination reaction reconstructed in a test tube
Rad51 recombinase forms a filament on a single-stranded DNA (substrate DNA1). Rad51 in the filament searches homologous duplex DNA (substrate DNA 2) and DNA strand-exchange reaction occurs once the recombinase finds homologous substrate.
(Figure below) Results of an investigation of the impact on the reaction when Fbh1 was added to the DNA recombination reaction mediated by Rad51 in a test tube
Fbh1 inhibited improper recombination reactions. This was observed as reduction in the amount of the final DNA product in the assay. On the other hand, when Fbh1 was added to proper recombination reaction timings, the amount of final DNA product increased, indicating that Fbh1 facilitates proper recombination reactions.
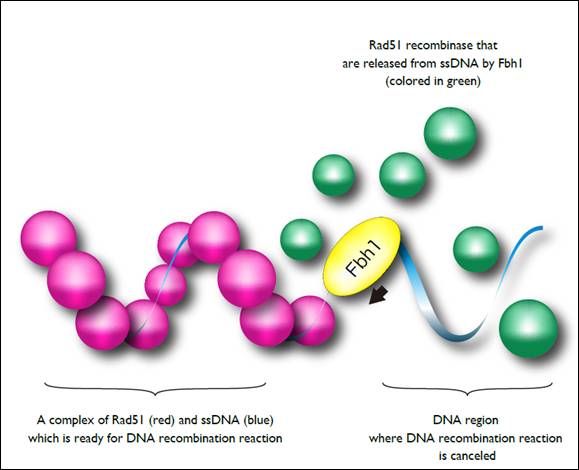
Figure 2. A molecular model of preventing improper recombination events by Fbh1
A filamentous complex formation of Rad51 recombinase and single-stranded DNA is an essential for homologous recombination. Fbh1 surveys the quality of Rad51-ssDNA complex, and disrupts the improper complex to protect initiation of the improper DNA recombination reaction.
Explanations of technical terms
|
Homologous recombination: Biological reaction that occurs with the exchange of two homologous DNA molecules (where the base sequences are very similar) and is involved in the repair of DNA double-strand breaks in mitosis. In meiosis, homologous recombination drives in shuffling genetic information of both maternal and paternal lineages (genetic information) to provide genetic diversity. It is also applied in the creation of genetically modified organisms. |
|
Fbh1: A DNA helicase widely conserved in organisms from fission yeast to human beings. Prototypes exist even in prokaryotes. |
|
Rad51: An enzyme that plays a central role in homologous recombination; it mediates searching for homologous DNA sequences and DNA strand exchange reactions. In prokaryotes, a protein called RecA has a similar function. |
|
DNA helicase activity: an enzymatic activity that unwinds double-stranded DNA into single-stranded DNA. |
|
Ubiquitin ligase activity: Ubiquitin is a small protein consisting of 76 amino acid residues, and is highly conserved among eukaryotic organisms. Through the cooperating of several enzymes, it attaches ubiquitin molecule to lysine residue in specific proteins (ubiquitination). Ubiquitin ligase activity is one of the essential enzyme activities for this reaction. Some ubiquitination are known to trigger protein degradation or transduce enzymatic activity of a target protein. |
Reference
Authors: |
Yasuhiro Tsutsui, Yumiko Kurokawa, Kentaro Ito, Md. Shahjahan P. Siddique, Yumiko Kawano, Fumiaki Yamao, Hiroshi Iwasaki |
Title of original paper: |
Multiple Regulation of Rad51-Mediated Homologous Recombination by Fission Yeast Fbh1 |
Journal: |
PLoS Genetics (2014) 10, e1004542
|
Digital Object Identifier (DOI): |
|
. Any information published on this site will be valid in relation to Science Tokyo.




