-Comprehensive analysis using a modified compound enabling free attachment/detachment of DNA-
Professor Toru Hisabori and Assistant Professor Satoshi Hara, of the Chemical Resources Laboratory at the Tokyo Institute of Technology, developed a new tool (marker compound) called "DNA-PC maleimide" that can simply and quantitatively detect the state of cysteine1, which is important for function/structure of protein in vivo. By using electrophoresis2 to examine proteins with this compound attached (conjugated), precise differences in the degrees of migration can be obtained. Furthermore, since the DNA moiety can be detached from the labeled protein using ultra-violet light, the redox state of the protein in vivo can be visualized using immunoblotting analysis3, which has widely increased the scope for applications in protein redox state analysis.
In recent years, the redox state of proteins in vivo has become an important factor in determining their physiological functions. Cysteine, which is one of the amino acids in protein, is particularly susceptible to the redox environment, and because it is also the key residue for intramolecular covalent bonds, which are important in the determination of protein structure, knowledge of its state has become important information in the clarification of protein function and its regulation.
Professor Hisabori and his team developed a compound called "DNA maleimide," which uses DNA as a chemical modifier, as a tool to probe redox states of cysteine. The compound only reacts to reduced cysteine and changes the molecular weight of proteins depending on the number of conjugated compounds, thus the state of the cysteine can be determined from the shift of migration in electrophoresis. However, because macromolecules like DNA are attached, it was unable to be used in immunoblotting analysis because DNA disturbs the analysis process. This drawback was resolved using a method enabling the easy removal of DNA moieties through exposure to ultra-violet rays, which then made it possible for the analysis.
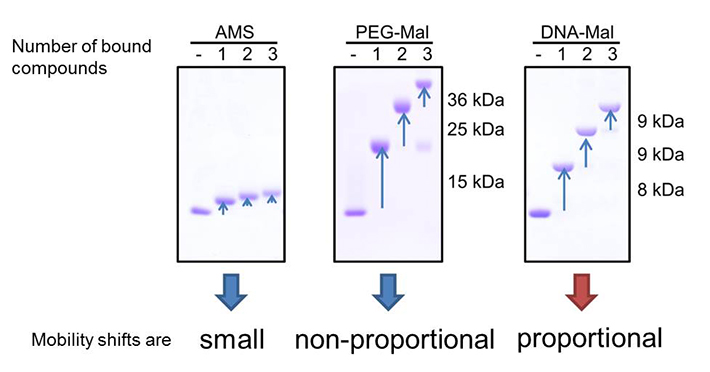
Comparison of mobility shifts derived from labeled maleimide compounds. AMS (left) derived mobility shifts are small. The mobility shifts of PEG-Mal labeled protein (middle) were not proportional and depending on the number of labeled maleimide compounds, making it difficult to determine the number of free thiols. In contrast, DNA-Mal (right) gave proportional mobility shifts, and the number of free thiols can be directly determined by the shifts.
Explanations of Technical Terms
1.Cysteine
One of the 20 kinds of amino acids that consist proteins; they have highly reactive thiol groups (SH groups) in their amino-acid side chains.
2.Electrophoresis
A technique that separates proteins in a fixed electric field. Proteins migrate and separate depending on its molecular weight.
3.Immunoblotting analysis
A technique that detects proteins separated by electrophoresis using an antibody that reacts to a specific protein (antigen). The proteins separated in the electrophoresis gel are transferred to a membrane made from nitrocellulose or similar, and an antigen-antibody interaction is induced on the surface of the membrane. Antibodies that bind with the specific protein are identified with another antibody (that has a coloring function) that reacts specifically to the primary antibody, enabling visualization.
Reference
Authors: |
Satoshi Hara, Yuki Tatenaka, Yuya Ohuchi and Toru Hisabori |
Title of original paper: |
Direct determination of the redox status of cysteine residues in proteins in vivo |
Journal: |
Biochemical and Biophysical Research Communications 2015 Jan 2; 456(1):339-43.
|
DOI: |
|
. Any information published on this site will be valid in relation to Science Tokyo.



