Scientists at Tokyo Institute of Technology uncover a sulfide-responsive protein that helps control photosynthesis in the purple bacterium Rhodobacter capsulatus.
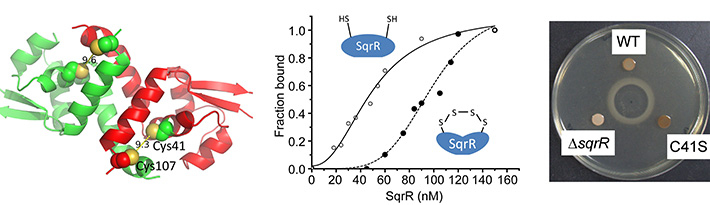
Figure 1. Action and function of the sulfide-responsive transcriptional repressor SqrR
Structural model of SqrR (left) showing sulfide-dependent tetrasulfide-bond formation that reduces its DNA-binding activity (middle). Mutation in sqrR influences photopigment synthesis (right).
Recent evidence demonstrates that the origins of photosynthesis can be found in deep sea hydrothermal vents, where microbes evolved to obtain energy from ejected hydrogen sulfide and methane gases. These microbes are capable of oxidizing sulfides and other gases and using them as electron donors to generate energy.
Unlike many micro-algae and plants that only use water as an electron donor to drive photosynthesis, purple bacteria such as Rhodobacter capsulatus can switch between energy sources (light or geothermal radiation) and different electron donors depending on their surrounding environment. To do this, the bacterium must carefully control the synthesis of electron transfer proteins in response to changing conditions. However, the precise mechanisms used by R. capsulatus to sense and make use of hydrogen sulfide are unclear.
Now, Takayuki Shimizu and Shinji Masuda at the Tokyo Institute of Technology, in collaboration with David P. Giedroc and Carl E. Bauer at Indiana University and researchers across Japan and the USA, have uncovered and characterized a sulfide-responsive protein, or transcriptional repressor, called SqrR, and described the mechanism by which it responds to sulfides. The findings shed light on electron donation processes in the early evolution of photosynthesis.
The team examined protein and gene responses in R. capsulatus. Genetic screening identified SqrR and showed that it acts as a sensor for reactive sulfide species inside cells. SqrR also regulates around 45% of the genes responsible for sulfide-dependent photosynthesis in R. capsulatus. It appears that, when sulfides in the surrounding environment increase, SqrR responds by binding to the sulfide molecules, thereby repressing photosynthetic electron transfer so that the bacterium can survive sulfide stress. In this way, SqrR helps maintain sulfide homeostasis in rapidly-changing environments.
Uncovering a major gene regulator like SqrR will allow scientists to examine photosynthesis in more depth, and determine how bacteria have evolved to survive in different environments. The findings may also have applications in synthetic biology.
Background
Bacterial photosynthesis
Purple bacteria have long been used by scientists to investigate the fundamental process of photosynthesis, precisely because they have adapted over millennia to respond rapidly to, and survive, multiple environmental stressors. The ability of bacteria such as R. capsulatus to use different electron donors — including hydrogen gas, methane and hydrogen sulfide — and switch between different radiation sources (light or geothermal) is of great interest to researchers, particularly those working in synthetic biology.
Advances in genetic screening now enable insights into the precise molecular mechanisms that control photosynthesis. While the enzymes involved in photosynthesis in purple bacteria have already been characterized, the work by Takayuki Shimizu and his team builds on understanding of how R. capsulatus senses sulfides in its surroundings and acts either to make use of, or protect itself from, the potentially toxic molecules.
By carefully monitoring and regulating the genes that facilitate electron transfer in sulfide-dependent photosynthesis, SqrR can help the bacterium survive sulfide stress. Further investigations will elucidate the processes that enable the bacterium to switch between different electron donors, and how components within R. capsulatus' cells are synchronised in response to their immediate surroundings.
Implications of the current study
The discovery of a major gene suppressor for R. capsulatus could enable further investigations into the early evolution of photosynthesis, and may also inform applications in synthetic biology.
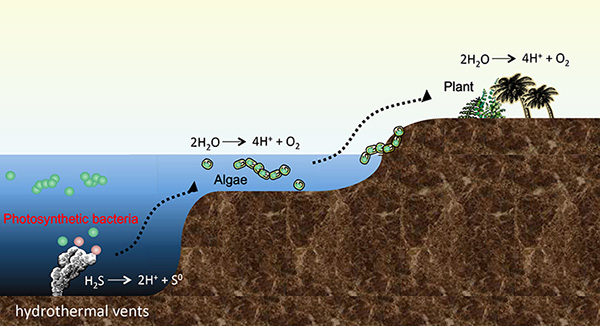
Figure 2. Model of the birth and evolution of photosynthetic organisms
Photosynthetic bacteria that were born in the deep sea hydrothermal vents have since evolved into algae, land plants. They were born for the first time on the Earth, it is thought that hydrogen sulfide (H2S) was used instead of water (H2O) as the electron source, that was their one or only one energy source.
Reference
Authors: |
T. Shimizu a, J. Shen b, c, M. Fang c, Y. Zhang b, d, K. Hori e, J. C. Trinidad b, d, C.E. Bauer c, D.P. Giedroc b, c, and S. Masuda f, g* |
Title of original paper: |
Sulfide-responsive transcriptional repressor SqrR functions as a master regulator of sulfide-dependent photosynthesis |
Journal: |
Proceedings of the National Academy of Sciences of the United States of America (PNAS) |
DOI : |
|
Affiliations : |
a Graduate School of Bioscience and Biotechnology, Tokyo Institute of Technology
b Department of Chemistry, Indiana University
c Department of Molecular and Cellular Biochemistry, Indiana University
d Laboratory for Biological Mass Spectrometry, Indiana University
e School of Life Science and Technology, Tokyo Institute of Technology
f Center for Biological Resources and Informatics, Tokyo Institute of Technology
g Earth-Life Science Institute, Tokyo Institute of Technology |
. Any information published on this site will be valid in relation to Science Tokyo.




