Scientists at Tokyo Tech, Nagasaki University have identified four potential protein inhibitors and unlocked drug discovery strategies for the treatment of Chagas disease by using advanced three-dimensional computer simulation by supercomputer TSUBAME in combination with in vitro experiments and X-ray crystallography. Through this "smart drug discovery" in which IT drug discovery and biochemical experiments cooperate, they identified hit compounds for target protein with a hit rate of 20 times or more than conventional High Throughput Screening (HTS) methods.
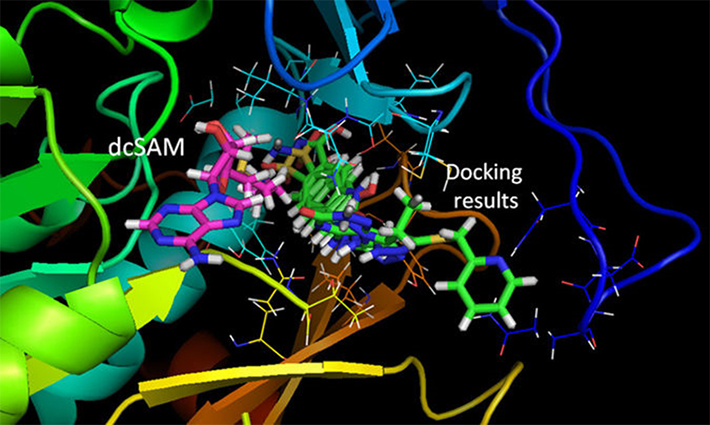
Figure 1.Results of the TcSpdSyn-ligand docking analysis.
This simulation was conducted with the T. cruzi SpdSyn X-ray structure (PDB ID code: 3BWC). The docking results show that all compounds bind to the putrescine-binding site and that dcSAM binding in its own site.
Chagas disease is a potentially life-threatening illness caused by the parasite, Trypanosoma cruzi, and is transmitted to humans through triatomine blood-sucking bugs that are commonly referred to as "kissing bugs" or "vampire bugs". While it was once confined to the Americas, worldwide travel has spread the disease, which is now endemic to approximately 20 countries. Currently, the World Health Organization (WHO) estimates that 10-13 million people are chronically infected, around 90 million people are exposed to the risk of the infection, and nearly 21,000 people die each year as a result, making effective treatment a necessity. Current treatments are largely effective in the first phase (acute) of the infection but have significantly diminished efficacy in the subsequent phase (chronic) of Chagas disease. Moreover, these drugs, which were developed in the 60s, are associated with severe adverse effects.
Masakazu Sekijima of Advanced Computational Drug Discovery Unit at Tokyo Tech, Kiyoshi Kita of School of Tropical Medicine and Global Health at Nagasaki University and -colleagues used a multi-modal integrated approach to develop potential new anti-Chagas therapies by combining the principles of structure-based drug design, where therapy is designed with the knowledge of the target's three-dimensional (3D) structure, with in vitro (colloquially referred to as "test tube experiments") testing methods, and X-ray crystallography. This approach narrows the range of potential drug candidates more efficiently. Through virtual screening by TSUBAME at Tokyo Tech, one of the world's top, large-scale supercomputers, they selected their target protein, T. cruzi spermidine synthase, based on specific structural features and properties indicating its importance for survival in another Trypanosoma species. If the protein is required for survival of a species, inhibiting that protein could be a potential mechanism of action for a drug with activity against the parasite that causes Chagas disease.
They focused on Spermidine synthase (SpdSyn) as the target protein, as sourced from the iNTRODB system. This system was developed by Prof. Yutaka Akiyama, Prof. Takashi Ishida of Department of Computer Science, at Tokyo Tech, and Prof. Kiyoshi Kita of School of Tropical Medicine and Global Health at Nagasaki University. Detailed information regarding the search method is presented in Figure 2. The in-house web-system iNTRODB facilitates the selection of drug target proteins for NTDs, particularly for trypanosomiasis. This system provides information on trypanosomal proteins with useful annotations, including the protein structure from the Protein Data Bank (PDB) and the protein inhibitors from ChEMBL.
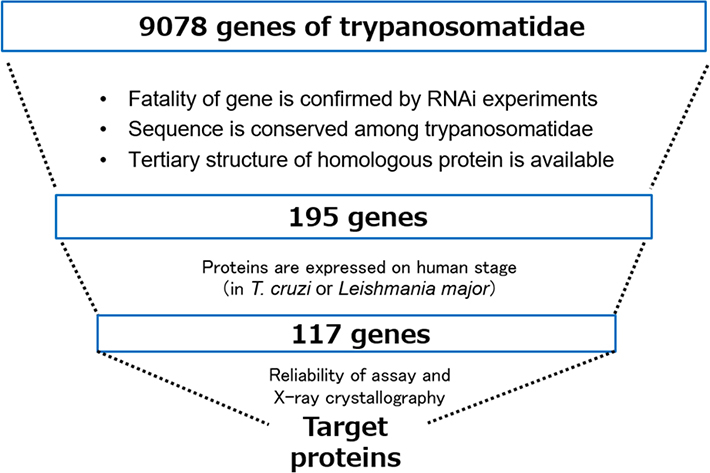
Figure 2.Search method for target proteins using iNTRODB.
Following selection, potential drug candidate inhibitors were identified through a screening search known as docking simulation-a structure-based drug design approach using 3D simulations to computationally match drug compounds to SpdSyn. They successfully identified four drug-like compounds that were virtual "matches" (referred to as "hits") then evaluated their inhibition activity in vitro and compared the results with those of a positive control. To further test potential activity and binding, they employed X-ray crystallography to confirm these four compounds in complex with the protein structure. Through interaction analyses for each compound the researchers found that all four compounds interacted with the proposed target binding sites through the same amino acid, Asp171. Additionally, molecular simulation suggested additional interacting sites for each compound that was not predicted by docking simulation.
Sekijima and team believe that their study's findings are indicative of the promise that docking simulation holds for the identification of potential drug-like inhibitors of the target protein and therapies for Chagas disease. They hope to demonstrate the general applicability of their approach, opening doors to the discovery of treatments for other diseases.
Reference
Authors : |
Ryunosuke Yoshino3,4,6, Nobuaki Yasuo2,6, Yohsuke Hagiwara2,5, Takashi Ishida1,2,6, Daniel Ken Inaoka7,8, Yasushi Amano5, Yukihiro Tateishi5, Kazuki Ohno2,5,9, Ichiji Namatame5, Tatsuya Niimi5, Masaya Orita5, Kiyoshi Kita7,8, Yutaka Akiyama1,2,6 , and Masakazu Sekijima1,2,3,6 |
Title of original paper : |
In silico, in vitro, X-ray crystallography, and integrated strategies for discovering spermidine synthase inhibitors for Chagas disease |
Journal : |
Scientific Reports |
DOI : |
|
Affiliations : |
1 Advanced Drug Discovery Unit, Institute of Innovative Research, Tokyo Institute of Technology
2 Education Academy of Computational Life Sciences (ACLS), Tokyo Institute of Technology
3 Global Scientific Information and Computing Center, Tokyo Institute of Technology
4 Graduate School of Agricultural and Life Sciences, The University of Tokyo
5 Medicinal Chemistry Research Labs, Drug Discovery Research, Astellas Pharma Inc.
6 Department of Computer Science, Graduate School of Information Science and Engineering, Tokyo Institute of Technology
7 Department of Biomedical Chemistry, Graduate School of Medicine, The University of Tokyo
8 School of Tropical Medicine and Global Health, Nagasaki University
|
. Any information published on this site will be valid in relation to Science Tokyo.





