Scientists unraveling the molecular details of DNA recombination regulation
Published: October 12, 2018
An international team from National Taiwan University (NTU) and Tokyo Tech identified how DNA recombinational repair is regulated in a recent work published in Proceedings of the National Academy of Sciences of the United States of America (PNAS). To repair damaged DNA, recombinases bind to DNA in order to initiate the repair work. However, the recombinase-DNA complex must be sufficiently stable to carry out the work. Scientists from NTU and Tokyo Tech work together to identify how another protein helps to stabilize this recombinase-DNA complex to efficiently proceed to proper DNA repair.
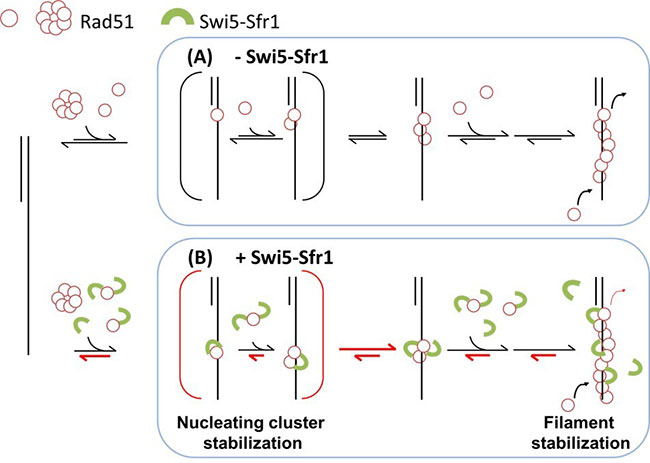
Figure 1. A molecular model for regulating Rad51 nucleoprotein filament formation by the Swi5-Sfr1 complex.
Swi5–Sfr1 stabilizes Rad51 on ssDNA primarily by preventing its dissociation. This stabilization effect leads to a stable nucleating cluster formation and a reduction in filament disassembly. Red half-arrows indicate the kinetic steps affected by SSwi5–Sfr1.
Prof. Peter Chi from NTU pointed out that "This work not only speaks to how assembly and disassembly of recombinase-DNA complex are regulated by this specific protein, but more importantly, it moonlights how other accessory proteins, such as BRCA2, the protein implicated in breast cancers, could affect filament stability and potentially derail the normal biochemical pathway in cancers."
"In addition to the beautiful science we did, this work stands as a model for the tremendous collaboration among biophysicists, biochemists and molecular biologists in Taiwan and Japan.", said Prof. Hiroshi Iwasaki in Tokyo Tech. The published work is only possible with the combination of different expertise in this international team.
This international team uses a special kind of tools, single-molecule imaging platforms, to watch how recombinases bind and dissociate onto individual DNA molecules with high resolution. "Watching the dynamics of these individual protein-DNA complex in real-time really offers information that is typically not available in conventional average-based experiments.", explained by Prof. Hung-Wen Li at NTU Chemistry Department. "Defects in DNA recombination are implicated in cancers. With this work, we now are in a position to design better strategies to battle DNA damages. This is just the beginning of the more exciting science in our joint adventure. It is fun and rewarding."
Figure 2. A microscope for single-molecule analyses used in this study.
This work "Swi5-Sfr1 Stimulates Rad51 Recombinase Filament Assembly by Modulating Rad51 Dissociation" is now in press in PNAS, and is carried out by Chih-Hao Lu, Hsin-Yi Yeh, Guan-Chin Su, Kentaro Ito, Yumiko Kurokawa, Hiroshi Iwasaki, Peter Chi, and Hung-Wen Li, where Iwasaki, Chi and Li are co-corresponding authors.
Reference
Authors : |
Chih-Hao Lua, Hsin-Yi Yehb, Guan-Chin Sub, Kentaro Itoc, Yumiko Kurokawac, Hiroshi Iwasakic, Peter Chib,d, Hung-Wen Lia |
Title of original paper : |
Swi5–Sfr1 stimulates Rad51 recombinase filament assembly by modulating Rad51 dissociation |
Journal : |
Proceedings of the National Academy of Sciences of the United States of America |
DOI : |
|
Affiliations : |
a Department of Chemistry, National Taiwan University
b Institute of Biochemical Sciences, National Taiwan University
c Institute of Innovative Research, Tokyo Institute of Technology
d Institute of Biological Chemistry, Academia Sinica
|
. Any information published on this site will be valid in relation to Science Tokyo.




