Scientists at Tokyo Institute of Technology (Tokyo Tech) , the Exploratory Research Center on Life and Living Systems (ExCELLS), and Nagoya University have succeeded in visualizing the binding processes of a class of cell-cell adhesion molecules called cadherins in solution at the nanometer scale with sub-second time resolution. They observed a novel conformation (S-shaped) of the cadherin dimer and the conformational transitions in the binding processes were revealed by single-molecule level observations. Direct visualization of the binding processes of cadherins at the single-molecule level will provide important insights into mechanisms of tissue- and organ-level organization, and diseases (e.g. cancer invasion) caused by cell-cell adhesion defects.
Cell, tissue, and organ structure is maintained by cell-cell adhesion molecules that connect opposing cells. Cadherins are a class of essential cell-cell adhesion molecules for tissue formation and integrity, and defects in cadherin function cause various diseases (e.g., cancer invasion). Cadherin protrudes from the cell surface and binds another cadherin on an opposing cell to mediate cell-cell adhesion. The cadherin binding process mainly comprises two dimerization steps: X-dimer formation and strand-swap (SS-) dimer formation of the extracellular domains (ectodomains) of cadherin. However, interactions other than those involving the formation of the X- and SS-dimers have also been proposed, and the precise binding mechanism of cadherin remains controversial.
Tadaomi Furuta of Tokyo Tech, Shigetaka Nishiguchi of ExCELLS and Takayuki Uchihashi of ExCELLS and Nagoya University applied high-speed atomic force microscopy (HS-AFM) to explore the binding mechanism of cadherins. HS-AFM can enable the visualization of single-molecule structures and dynamics in solution at the nanometer scale with sub-second time resolution by directly touching and scanning the surface of proteins through a sharp-tipped probe. HS-AFM revealed that cadherins existed as multiple dimeric structures, which based on their morphology may be classified as W-, cross-, and S-shaped dimers. Furthermore, the scientists conducted mutational and structural modeling analyses and found that W- and cross-shaped dimers corresponded to known SS-dimers and X-like dimers and that the S-shaped dimer is a novel conformation. The binding processes of cadherins directly visualized by HS-AFM also revealed that the dimerization process is completed within 1 second through conversion into the aforementioned three types of dimeric structures. Based on these HS-AFM observations, the scientists hypothesized that the binding mechanism progress through the sliding motion of the S-shaped dimer followed by the flipping motion of the X-dimer to form the SS-dimer, which is thought to be the final stable cadherin dimer (Figure 1).
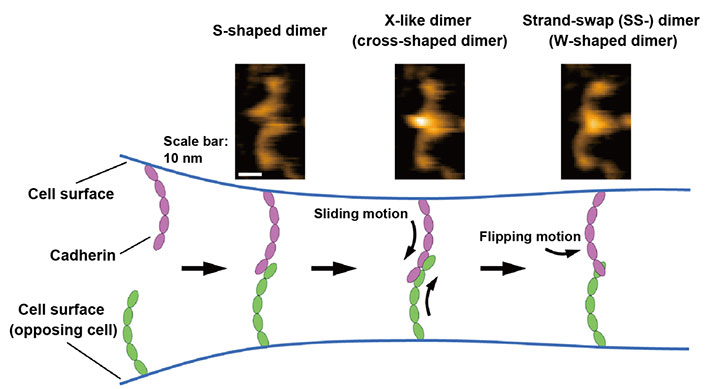
Figure 1. Binding processes of cadherins revealed by high-speed atomic force microscopy (HS-AFM).
HS-AFM images of the cadherin dimers are shown at the top. The binding mechanism of cadherins is illustrated at the bottom based on HS-AFM observations.
To date, the binding mechanism of cadherins has been mainly investigated using structural analyses and cell and solution measurements, which can only analyze the binding states reflected by the large number of cadherins. The newly applied HS-AFM technique revealed the binding processes of individual cadherins at single-molecule resolution, which has not been achieved before. HS-AFM observation will pave the way for a deeper understanding of the binding mechanism of cadherins, which is important for tissue- and organ-level organization and cell-cell adhesion-related diseases.
This work was supported by the Foundation of Public Interest of Tatematsu (S.N.). This work was supported in part by the Grants-in-Aid for Scientific Research (grant numbers: JP 21H01772 and JP 21H00393 [T.U.]) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan. This work was supported by the Joint Research of the Exploratory Research Center on Life and Living Systems (ExCELLS; program no. 18-101 [T.U.]).
Reference
Authors : |
Shigetaka Nishiguchi1, Tadaomi Furuta2, Takayuki Uchihashi1,3,4 |
Title of original paper : |
Multiple dimeric structures and strand-swap dimerization of E-cadherin in solution visualized by high-speed atomic force microscopy |
Journal : |
Proceedings of the National Academy of Sciences of the United States of America |
DOI : |
|
Affiliations : |
1 Department of Creative Research, Exploratory Research Center on Life and Living Systems, National Institutes of Natural Sciences, Okazaki, Japan
2 School of Life Science and Technology, Tokyo Institute of Technology, Yokohama, Japan
3 Department of Physics, Nagoya University, Nagoya, Japan
4 Institute for Glyco-core Research (iGCORE), Nagoya University, Nagoya, Japan
|
- *
- This article was updated on July 25 to add DOI.
. Any information published on this site will be valid in relation to Science Tokyo.