Technology that creates the world
"Genome editing technology" edits the genome (genetic information)[1], which is the biological blueprint, to change the traits of an organism as desired. It has recently been in the global spotlight due to its wide-ranging fields of application. Professor Yuriko Osakabe engages in fundamental research for genome editing technology and developed a novel domestic genome editing tool. Welcome to a world of dramatically advancing technology that unlocks new possibilities for life.
Revolutionary genome editing tool "TiD"
All living organisms have a genome (although there are differences in the number of genes it contains) which determines their appearance, physiological characteristics, and biology. Genome editing technology induces the expression of different traits by changing only the targeted DNA of the genome. Two researchers who worked on the current mainstream genome editing tool "CRISPR-Cas9"[2] won the Nobel Prize in Chemistry in 2020.
My research in the field of genetic engineering has progressed along with changes in genome editing technology. I started my research in the 1990s, when the term "genome editing" did not even exist. Subsequently, the development of editing tools progressed rapidly around the world, and CRISPR-Cas9 appeared in the 2010s. As a result of our vigorous efforts to develop and demonstrate a novel method, we finally developed the world's first novel genome editing tool "TiD" and successfully applied for a patent in August 2018. What makes TiD special is that the incidence of "off-target effects" in which the base sequence[3] of non-target DNA is edited (which was an issue with CRISPR-Cas9) is extremely low. My current focus is detailed research aimed at the practical application of TiD.
Possibilities for genome editing technology via industrial applications
"We will clarify one by one why organisms live and why they have a variety of functions, and find clues to solving social problems. It is a field of research where you truly feel that your own discoveries will contribute to the world."
In 1992, completed a master's degree at Graduate School of Agriculture, the University of Tokyo,. In 1996, completed a doctoral degree at the United Graduate School of Agricultural Science, Tokyo University of Agriculture and Technology. Doctor of Agricultural Science (Tokyo University of Agriculture and Technology). Involved in the development of TiD as part of the smart cell project by the New Energy and Industrial Technology Development Organization (NEDO).
TiD has expanded the applications of genome editing technology in a major way. This is because the high accuracy of editing will lead to the adoption of genome editing technology in medicine and drug discovery, which has been challenging in terms of safety. If TiD can be put to practical use, it will provide a hopeful prospect for the treatment of serious genetic diseases and intractable diseases such as cancer, and cell therapies. Applications for iPS cells and virus detection are also envisioned. Of course, it can also be applied to other familiar industries. For example, its adoption in the field of reusable bioresource development made from plants and microorganisms could greatly improve production efficiency and solve the energy problem. Using it for breed improvement in the agricultural and livestock industry could solve food problems by developing breeds with high nutritional value, acquiring resistance to environmental stress, and increasing yields. Genome editing technology (including TiD, which is now in the global spotlight) has the potential to produce solutions to complex social problems.
Applications in medicine
The pinpoint repair of faulty genes will likely lead to gene therapies for hereditary diseases, cell therapies to edit patient cells, and the discovery of novel cancer treatments.
Applications for drug discovery
In addition to helping to streamline the drug-making process, applications have also been found that improve the safety of vaccines administered to humans and animals.
Applications for bioresource development
Enabling the production of cells with advanced functional design has also promoted the increased production of bioresources and the development of high added value resources.
Applications in agriculture, livestock, and fisheries
Its use in breeding has led to the development of highly nutritious agricultural and livestock products, large farmed fish that can help solve food problems, and plants that can grow in harsh environments.
Old friends sometimes ask me if research is a struggle, but for me the fun always wins out over the hardships. The experiments involve plants so perseverance is required, but once you have experienced the excitement of the moment when your hypothesis is proven, there will be no stopping you. I will never forget the thrill of being a researcher who unravels the unexplained, and will continue to explore the principles of life.
Decipher and use "biological blueprint"

Organisms all have a species-specific genome. For a long time, humankind has puzzled over how to decipher and use this "biological blueprint."
A genome is the set of genes that an organism has. In 2003, the human genome was successfully deciphered after 13 years of work, and the genome decipherment of many organisms is still underway.
"Once we learned about the base sequences that make up the DNA of a genome and the functions of genes, the idea arose to see if we could use that information to overcome existing problems or make better use of it. That is the origin of genome editing, which changes the function of genes." (Osakabe)
Genome editing is a technology that "edits the base sequence of DNA and changes the functions of genes precisely as intended." The principle behind genome editing is to first cause a DNA double-strand break at a specific part with an artificial enzyme, nuclease, also called "molecular scissors." Then, mutations caused by repair errors that occur when organisms repair the break are used to change traits. In fact, mutations themselves are a frequently occurring phenomenon in the natural world, and genome editing can be visualized as artificially flipping a switch to make it happen. In other words, this technology is probably close to a natural law.
The history of genome editing technology is the trajectory of the development of artificial nucleases, that is, enzymes that are artificially modified to degrade DNA, and the development of technologies that optimize these genome editing tools for application in living organisms. Early genome editing tools, ZFNs, were developed in the late 1990s, followed by TALENs, and in 2012, CRISPR-Cas9 (used today as a standard worldwide) was first reported.
Genome editing as a "DNA repair mechanism"
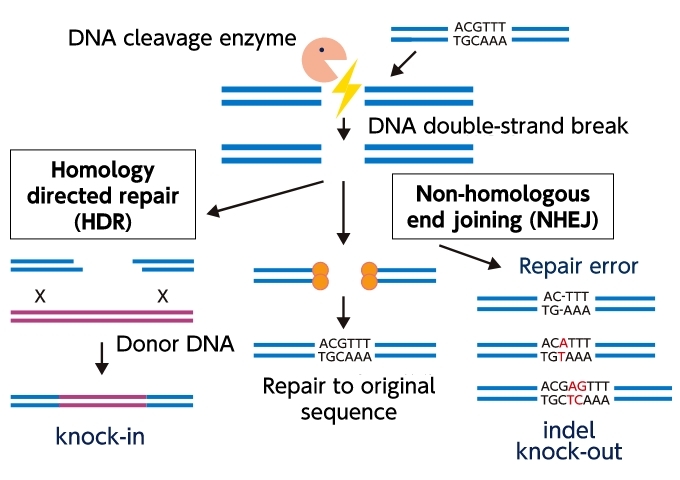
Genome editing employs the method of introducing mutations (knock-out) by utilizing the fact that three patterns of repair errors (indels) occur around the site of DNA double-strand breakage ((1) deletion of base, (2) substitution with another base, and (3) insertion of another base). Alternatively, scientists use the method of rewriting a DNA sequence by inserting donor DNA and copying the base sequence during repair (knock-in).
Researchers around the world have accepted the challenge of developing genome editing tools that are more accurate and easier to use, and new ones have been developed one after another. CRISPR-Cas9 (currently the mainstream tool) is easy to produce and anyone can use it for fundamental research, so the possibilities of applying genome editing in various fields have suddenly expanded, a so-called "biotech revolution."
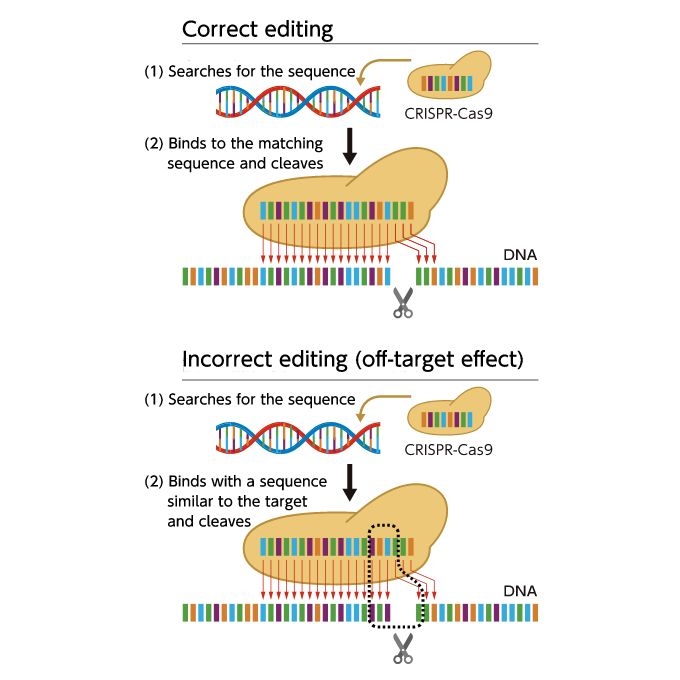
But there are still hurdles to cross. Genome editing using CRISPR-Cas9 carries the risk of editing unintended genes. CRISPR-Cas9 identifies the target base sequence, binds to that part, and cleaves the DNA. CRISPR-Cas9 may mistakenly bind to or cleave parts of a similar base sequence existing in the genome, resulting in the introduction of mutations. In that situation, there are cases where the gene does not work as desired, and it becomes necessary to remove these mutations. This is an "off-target effect" and is one of the challenges to overcome in order to utilize genome editing in the fields of medicine and drug discovery.
The world's first novel genome editing tool that can significantly mitigate this off-target effect is TiD, developed by Osakabe and others. TiD has an order of magnitude lower risk of editing unintended genes than conventional tools. The reason is that it identifies longer sequences than CRISPR-Cas9. CRISPR-Cas9 uses a 20-base sequence, whereas TiD uses a 35- or 36-base sequence to identify edit sites, so the probability of a similar sequence existing in the genome is very low. This allows for precise target identification and reduces the risk of off-target mutations and deletions. Because of this unique feature, TiD has paved the path for genome editing to expand into medicine and drug discovery, among other industrial applications.
Most existing genome editing technologies, including CRISPR-Cas9, were developed in Europe and the United States, and their commercial use in Japan raises issues with intellectual property rights. Since TiD is domestically produced, it is also attractive for reducing the costs of using foreign patents. The real-world implementation of TiD will undoubtedly serve as a foothold for the promotion of an independent Japanese industry that applies genome editing.
Issues at hand "Off-target effects"
CRISPR-Cas9 searches for and binds to 20 specific bases out of hundreds of millions of base pairs in living organisms. However, if there are similar sequences in the genome that differ by only one or two out of the 20 bases, it may bind incorrectly. This is the "off-target effect."
The real-world implementation of genome editing technology requires not only optimization of technology according to individual organisms, but also cell regeneration (to demonstrate normal expression of desired traits in edited organisms) and peripheral technologies (such as genome analysis to determine where and how to edit in the base sequence of the target organism).
For example, in the world of plants (one of my research subjects), there is still much to do in genome analysis of various plant species and cultivars, and many issues remain such as there being species for which tissue culture and plant regeneration have not succeeded over many years. Genetic engineering researchers must also tackle these issues. We are accumulating interdisciplinary knowledge with researchers from other fields, which should lead to new achievements in the near future. (Osakabe)
TiD is expected to change the landscape of Japanese genome editing technology, and research is currently underway with the aim of real-world implementation in collaboration with Tokushima University (which is jointly developing the technology) and companies conducting various research using TiD technology. Osakabe is pouring effort into this.
"We are striving daily for the practical application of TiD, which has potential in various fields such as medicine, drug discovery, bioresource development, and improvement of food species. We would like to get the necessary basic technologies ready so that we can contribute to solving social problems where complex fields are intertwined."
Genome-editing technology holds great power. Researchers will continue to discuss how to use it properly, and devise methods that will be useful to people and society. The young genetic engineering researchers at Osakabe Laboratory are advancing their research step by step, and at the same time gradually realizing the social significance of their fundamental research. Dreaming of the day when we unravel the mysteries of life and design new forms of life.
Terms
[1]
Genome (genetic information) : The genetic information of all DNA (deoxyribonucleic acid) contained in an organism, which shapes the organism and is necessary for life activities. The information in the DNA of a genome that specifies the order of amino acids when making a protein for a living organism is called a gene.
[2]
CRISPR-Cas9 : A genome editing tool that uses the immune system of bacteria to cleave DNA double-strands and introduce mutations at arbitrary sites in DNA. It is characterized by using RNA molecules to identify DNA sequences, and has the advantage of being simpler and less costly than conventional tools such as ZFNs and TALENs.
[3]
Base sequence : DNA forms a double helix structure by pairing at the bases of nucleotide molecules. Genetic information is represented by a sequence composed of four base types, A (adenine), T (thymine), G (guanine), and C (cytosine).
. Any information published on this site will be valid in relation to Science Tokyo.