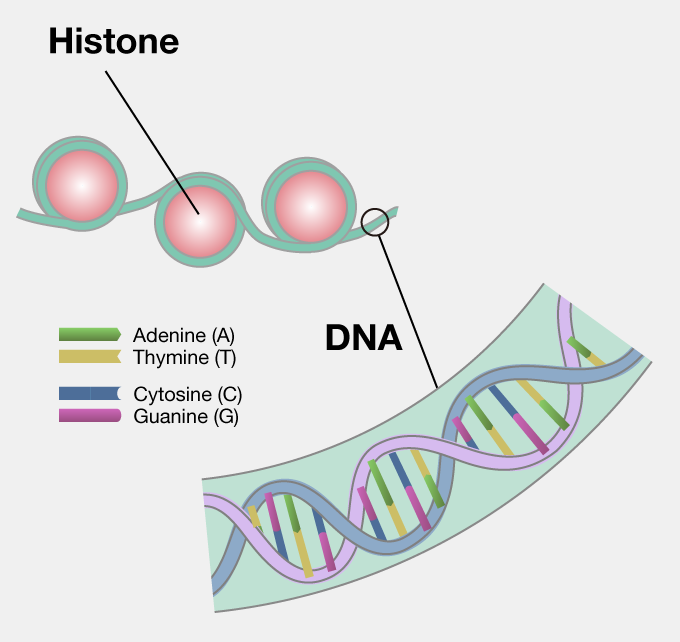
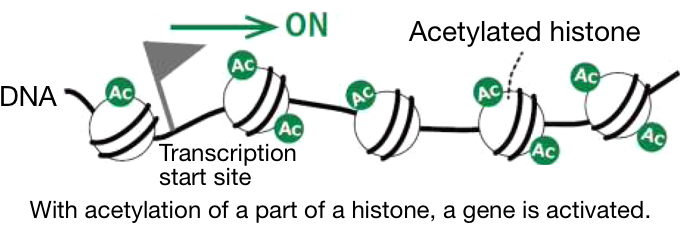
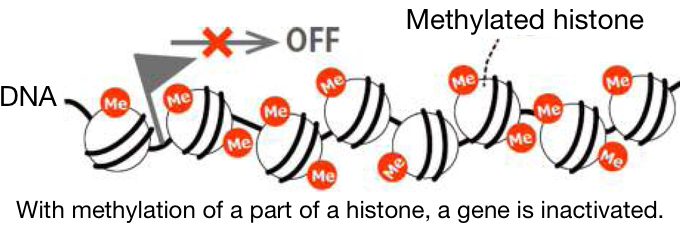
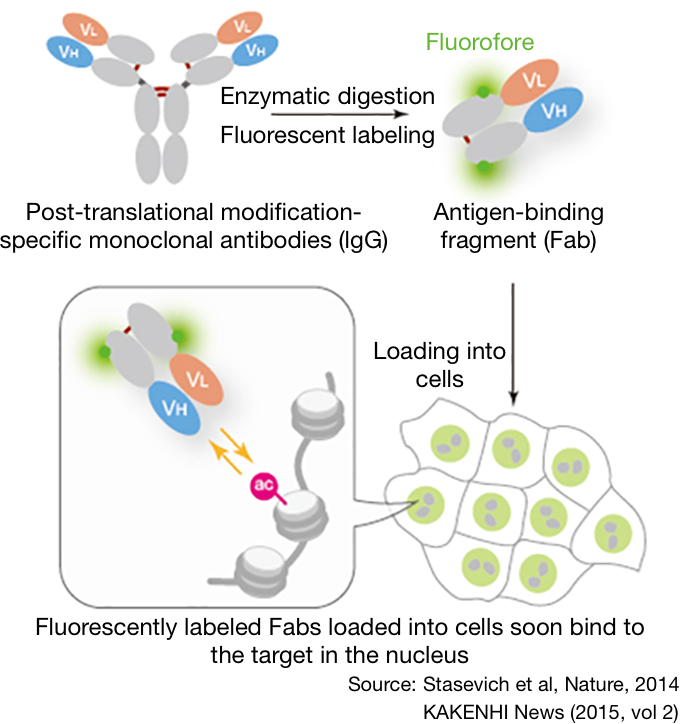
1 Atom groups
Groups of atoms have covalent bonds in molecules of compounds and can move from one molecule to another. They are the same as the functional groups within molecules such as the methyl group (CH3-), ethyl group (C2H5-), acetyl group (CH3CO-), hydroxyl group (HO-), and the phosphate group (H2PO4-). They are usually connected to other atoms within molecules.
2 Stem cells
The human body continuously creates cells to maintain health. Stem cells act as a supply source for the cells. There is a wide variety of stem cells, each thought to form different organs. Hemopoietic stem cells, for example, are responsible for creating blood cells. Embryonic stem cells can become any tissue or organ such as blood, bones, skin, and brain. In addition, iPS cells are cells artificially made multipotent and which have nearly the same characteristics as embryonic stem cells.
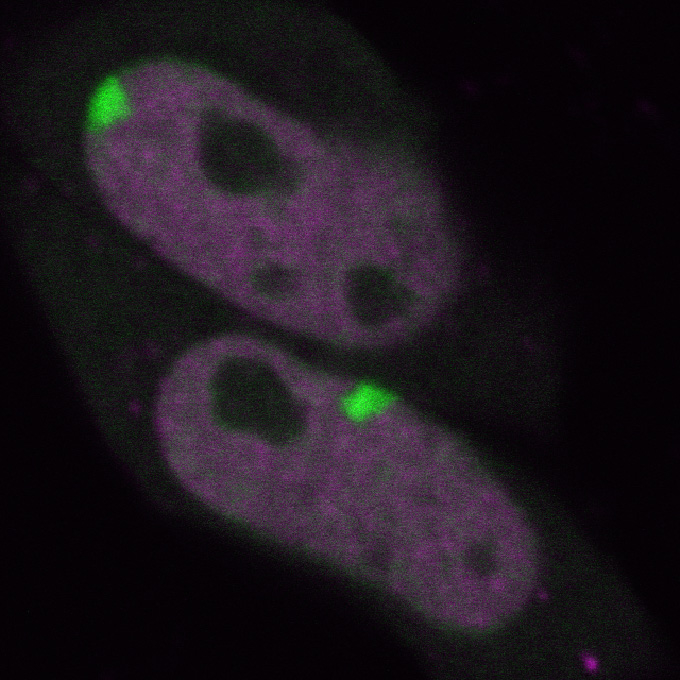
3 Fluorescent proteins
Fluorescent proteins exhibit fluorescence. Any protein can be genetically fused with a fluorescent protein for expression in living cells, which allows us to reveal the location and movement of protein molecules. Osamu Shimomura, the 2008 Nobel Prize in Chemistry laureate, purified green fluorescent protein derived from jellyfish Aequorea victoria.
4 Central dogma
The central dogma is the basic principle of molecular biology developed by Francis Crick, one of the scientists that discovered the double helix structure of DNA in 1958. The central dogma explains that genetic information in all living creatures is passed from genome DNA (copied), transcribed into mRNA, and then translated into proteins. However, the discovery of synthesis of DNA from RNA in 1970 and other discoveries have modified this original concept. Attempts to clarify the concept of the central dogma remain very meaningful, though, as advanced studies on gene expression have led to various discoveries, such as mRNA and tRNA.
. Any information published on this site will be valid in relation to Science Tokyo.