Our bodies are equipped with amazing biological systems such as immunity and metabolism. These systems recognize our surrounding conditions, process information, and operate autonomously. But despite these systems being composed entirely of known substances, we have yet to be able to artificially create living systems. Where is the boundary between matter and life? Professor Masahiro Takinoue (Department of Computer Science, School of Computing) is working to solve this mystery from the perspectives of both physics and life sciences. Professor Takinoue is engaged in research using artificially synthesized DNA to create artificial cells and molecular robots that function autonomously.
Artificial cells capable of autonomously processing and storing information
- Tell us about your research.
I'd be happy to. To start with an example, the human body has a function called immunity. Immunity maintains our health. Cells engage in a series of behaviors which consist of detecting the intrusion of an enemy such as a virus or bacteria, identifying the intruder as an enemy, and then pursuing and attacking the intruder. When viewed from the perspective of physics and engineering, this behavior is akin to an autonomous robot that combines information processing and motor functions. Moreover, the energy to drive this cellular behavior is obtained from sources such as the surrounding blood. The cells operate using an extremely low amount of energy. All of these cells are composed of substances that can be obtained by researchers. If we were able to artificially manufacture such cellular behavior, it would be useful for the prevention and treatment of diseases. Furthermore, autonomous robots have a wide range of applications. For example, in addition to being useful for exploration in the human body, they can explore the deep sea, outer space, and other areas which are difficult for humans to access.

Therefore, our laboratory is conducting research on artificial cells and molecular robots1 Researchers in various countries around the world are also conducting research on artificial cells. However, a major distinguishing feature of our laboratory is that we use artificially synthesized DNA2 as a material.
My specialty is physics and information science, not biology. Consequently, I am most interested in how to combine substances to build complex autonomous systems like cells. Therefore, my research theme is to artificially create different types of living systems and connect them to engineering applications, while also pursuing the operating principles of living systems.
- How do you make artificial cells and molecular robots using DNA?
There are two main reasons that we use DNA. The first is the ability to incorporate dynamic information processing, and the second is the ability to create a three-dimensional structure in accordance with design through self-assembly. Self-assembly means that atoms and molecules gather to form a naturally stable structure, similar to how water molecules gather to form a snowflake.
The first important point is that, DNA has four types of bases: adenine (A), thymine (T), guanine (G), and cytosine (C). Base pairs formed by A-T and G-C create a double helix structure of DNA and store genetic information. In other words, the foremost feature of DNA is that information can be incorporated into the molecule by specifying the base sequence. This feature cannot be found in other substances, such as plastic.

Using properties like this, we can create a molecular computer. By skillfully designing base sequences, we can perform computations and store information. By installing molecular computers in artificial cells or molecular robots, we can give them autonomy. Originally, this field was pioneered by Professor Leonard Adleman of the University of Southern California about 30 years ago. However, the pursuit of how to process information with molecules is still an important research theme. For molecular computers, we will build completely new calculation methods that do not exist in conventional computers.
The second point is that by designing the base sequence of DNA we can freely create various three-dimensional structures other than the double helix structure. Professor Nadrian Seeman of New York University invented a technique for creating arbitrary two-dimensional or three-dimensional nanometer-sized structures (nano is on the scale of 10-9). We can use electronic computers to predict what kind of base sequence can be used to create a certain three-dimensional structure. Simply by mixing the DNA with the designed base sequences in a solution and incubating it for a certain period of time, the DNA will self-assemble and create the desired three-dimensional structure.
Our laboratory is located within the School of Computing, which mainly conducts research on electronic computers. However, we possess research facilities for not only electronic computers but also biophysical wet experiments; I believe that such an interdisciplinary environment will create new science fields.

Mix the DNA in solution (left) (above) and place it in a dedicated device for a certain period of time (right) (below).
The desired three-dimensional structure is achieved through self-assembly.
An artificial cell by accumulation of DNA origami
- Tell us in detail about your research themes.

I am currently conducting research on two main themes. The first is research on cell-sized molecular robots shaped like capsules.
Our goal here is to create cell-like molecular robots from DNA and proteins with specific functions, such as exterminating viruses and cancer cells. Multiple molecular robots would then exchange information and operate in cooperation like an immune system.
In order for molecular robots to cooperate with each other, it is necessary to equip the membranes that compose the capsule with an information exchange function. Therefore, we make the membrane from DNA. By making the membrane function as an interface with its surroundings, we enable the capsule to move autonomously in response to external stimuli. In addition, the human body's immune system stores information about enemies such as viruses. By using DNA in the membrane, we also equip the molecular robot with the ability to store information.
My second research theme is to produce artificial cells using microchannels. The maximum size of a molecular robot that can be manufactured through self-assembly is 100 nanometers. This is about 1/1,000 of the thickness of hair. We need different techniques to make larger molecular robots and artificial cells. Therefore, our laboratory has used microchannels. We can manipulate DNA by flowing a solution containing DNA through the microchannel. By using this process, we can create larger molecular robots and artificial cells.
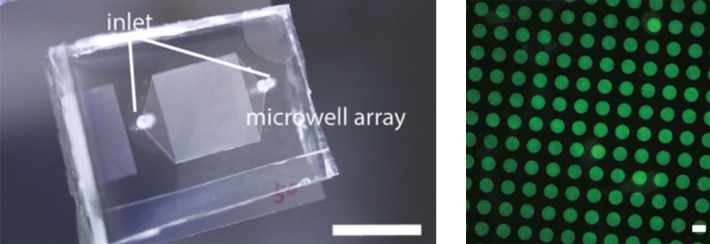
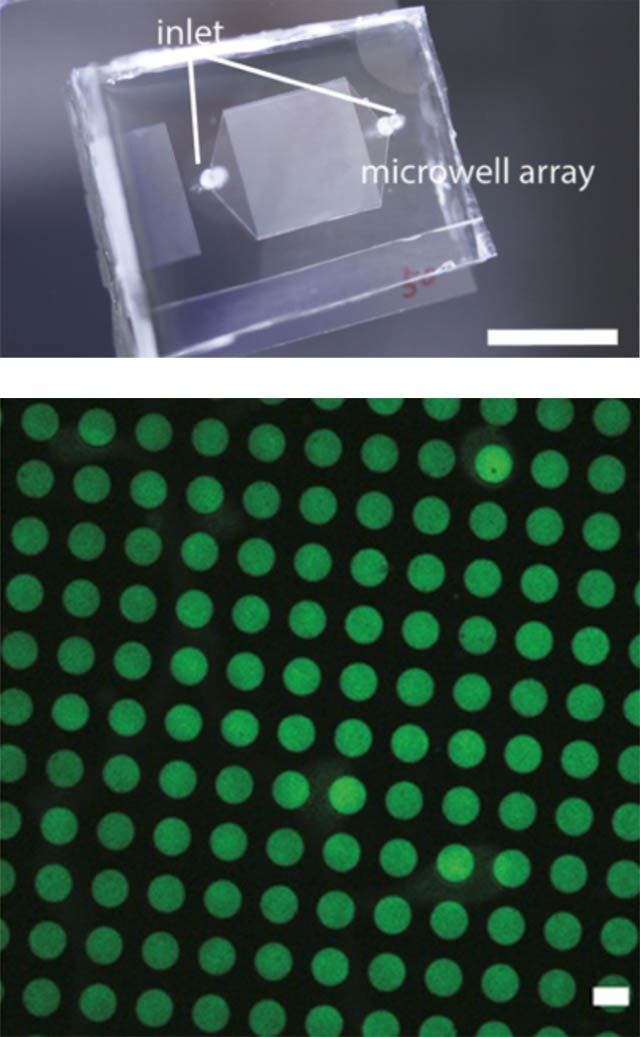
External view of a microchannel (left) (above). A microchannel is a plate in which micrometer-wide fine channels and wells (right) (below) are formed in a thin substrate made of transparent silicone rubber and glass. By flowing a solution containing DNA through the microchannel, we can produce molecular robots and artificial cells of a large size.
- What are your main findings so far?
In 2016, we discovered a phenomenon in which DNA gel behaves like a liquid in certain conditions, and in which separate DNA gels fuse together and change shape. Until now, DNA gels were thought to maintain a constant shape, so it was a huge surprise when we saw the gel in liquid form.
Moreover, we also found that simply changing a few base sequences of the DNA significantly changes its properties. The information of only a few bases controlled the properties of a gel with a cell size 1,000 times larger than the bases. We are currently developing this technology into computational DNA droplets that detect microRNAs (miRNAs), which are biomarkers for cancer.*
- *
-
Novel Method for Early Disease Detection Using DNA Droplets | Tokyo Tech News (Published: June 6, 2022)

By designing the base sequences of DNA, it is possible to prevent DNA gels from fusing together. In the image, DNA gels of like colors fuse together (green to green, blue to blue), but DNA gels of different colors do not. This was achieved by changing just a few base sequences.
Another major result recently made by our laboratory was the creation of artificial cells by a membrane-like structure made of a large accumulation of DNA origami3. I was thrilled when I used a fluorescence microscope to confirm that DNA origami accumulated and made capsules by self-assembly.
The DNA origami has a structure with a pore in the center, so the surface of the capsule also has multiple pores. We are now able to achieve a communication function by releasing a substance from the pore and exchanging the substance between capsules.
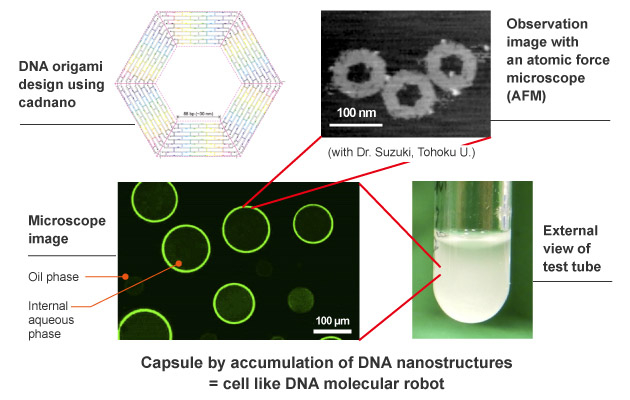
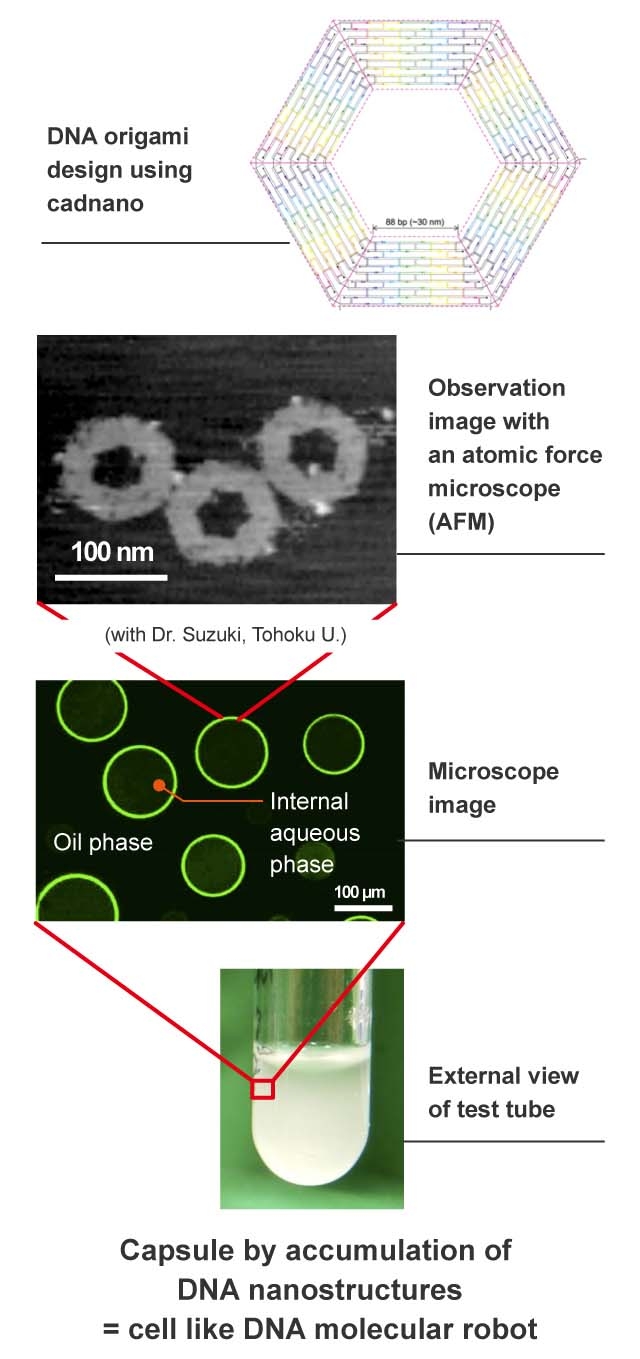
DNA origami has a hexagonal shape in which DNA is folded by self-assembly. We create capsules by having them overlap by themselves through the process of self-assembly. The diameter of the DNA origami is 100 nanometers, and the diameter of the capsule is 100 micrometers.

Concept image of artificial cell microcapsules formed using DNA origami (Ishikawa, et al., Angew. Chem. Int. Ed. (2019))
The fundamental differences between matter and life
- What led you to starting your research?
When I was in high school, I read an article in a scientific magazine about "Conway's Game of Life," which was devised by Professor John Horton Conway of Princeton University. The article gave me the desire to study artificial life. Conway's Game of Life is a simulation that imitates the process of life; for example, birth, evolution, and selection. Ever since I was a kid, I had wondered what it meant to be "alive." When I learned about Conway's Game of Life, I was excited at the potential for grasping the essence of living things through mathematics and physics.
On the other hand, ever since I was in junior high school, I had a vague desire to become a researcher. At the same time, I was fascinated by Albert Einstein's theory of relativity and enjoyed physics. At the time, I was wondering how to balance both of my interests. Then, upon entering university, I learned that there was a field of biophysics where I could study both biological systems and physics. I knew that this was my calling in life. I decided to enter the Department of Physics and study biological systems from the perspectives of physics and information science.
However, although the concept of Conway's Game of Life is extremely interesting, I was worried that it had no material substance. Consequently, I began to feel the desire to create artificial cells using actual molecules. Then, upon entering graduate school, I learned that the laboratory run by my supervisor Professor Akira Suyama (Professor Emeritus at the University of Tokyo) was researching molecular computers using DNA, and I decided on molecular computers as the theme of my doctoral dissertation.
- Tell us about your future goals.
My short-term goal is to enable artificial cells and molecular robots to perform complex information processing using molecular computers. My long-term goal is to link artificial cells and molecular robots to each other, thereby achieving systems that can perform important functions, similar to the immune system.
My ultimate goal is to examine the fundamental theme of differences between matter and life. Since biological systems are actually composed of non-living matter, our own bodies have demonstrated that living systems can be created using non-living matter. However, the reason why we have not been able to artificially create living systems is that doing so requires understanding physical and chemical design principles of living systems that we humans do not yet know. The ultimate purpose of my research is to seek out the design principles, which are something that I find very interesting.
The research that we conduct can be quite challenging, and we often face completely unexpected results from our experiments. The basis of our research is not only to create artificial cells and molecular robots, but also to investigate the mathematics and physics of the biological systems behind them. Therefore, when we encounter a physical or chemical phenomenon that differs from our imagination, asking the question "Why?" is always a new starting point for research.
The thrill of being a researcher is to encounter the unknown
- What message do you have for students who want to become researchers?

For me, the real thrill of being a researcher is the moment when I encounter something that is unknown to anyone else in the world.
My advice for young people who aspire to become scientists is to devote their interests to any subject that attracts them. Research is not only fun; it is also difficult. Therefore, I advise young people to pursue their true interests, without being overwhelmed by the opinions of others and the trends of the times. Devoting yourself to the pursuit of your interests will open up new paths which you never expected.
1 Molecular robot
An artificial molecular system that integrates molecular devices (sensors, computers, actuators, etc., designed using molecules). By receiving external molecular signals and making decisions through molecular computing, molecular robots possess the senses, intelligence, and movements for reacting autonomously to the environment.
2 DNA
DNA is an abbreviation for deoxyribonucleic acid. DNA possesses advanced molecular recognition ability based on the sequence information of four types of bases, A, T, G, and C, and is responsible for the storage and transmission of genetic information in a living organism. In recent years, the chemical synthesis of DNA has become easier. By using its molecular recognition ability, DNA can be used to record complex nanostructures (e.g., DNA origami) and digital data. It is also applied to technology such as DNA computers that are capable of solving mathematical problems.
3 DNA origami
Two-dimensional or three-dimensional DNA nanostructures composed of a long single-stranded DNA (approximately 7,000 bases) and many short single-stranded DNA (several dozen bases). By folding a long single-stranded DNA into a single stroke according to our desired shape and fixing it with short single-stranded DNA designed to complement each other, a structure that is several dozens of nanometers in size can be created. Reported in 2006 by Dr. Paul Rothemund of the California Institute of Technology.

Masahiro Takinoue
Department of Computer Science, School of Computing
- April 2022 -Professor, School of Computing, Tokyo Institute of Technology
- 2016 - 2022Associate Professor, School of Computing, Tokyo Institute of Technology
- 2011 - 2016Associate Professor, Interdisciplinary Graduate School of Science and Engineering, Tokyo Institute of Technology
- 2009 - 2011Project Assistant Professor, Institute of Industrial Science, The University of Tokyo
- 2009 - 2009Postdoctoral fellow, Institute of Industrial Science, The University of Tokyo
- 2008 - 2009Postdoctoral fellow, Division of Physics and Astronomy, Graduate School of Science, Kyoto University
- 2007 - 2008JSPS Research Fellowship for Young Scientists (PD), Graduate School of Arts and Sciences, The University of Tokyo
- 2007Ph.D. in Physics, Department of Physics, Graduate School of Science, The University of Tokyo
- 2004M.Sc., Department of Physics, Graduate School of Science, The University of Tokyo
- 2002B.Sc., Department of Physics, Faculty of Science, The University of Tokyo

NEXT generation is a new series about emerging researchers, working at the forefront of science and technology as they envision the impact their discoveries will have on society's future.
The Special Topics component of the Tokyo Tech Website shines a spotlight on recent developments in research and education, achievements of its community members, and special events and news from the Institute.
Past features can be viewed in the Special Topics Gallery.
. Any information published on this site will be valid in relation to Science Tokyo.


























